Recently, the Laboratory of Structural Biochemistry at SIAIS, led by Distinguished Adjunct Professor Roger Kornberg and Research Associate Professor Zhang Heqiao, published a research article entitled “Class I histone deacetylase complex: structure and functional correlates” in the Proceedings of the National Academy of the Sciences (PNAS), reporting a 3.2 Å resolution cryo-EM structure of the class I histone deacetylase (HDAC) complex.

The nucleosome is the fundamental unit of chromosomes in eukaryotes. The 147-base pair DNA wraps around the histone octamer, comprised of H2A, H2B, H3 and H4, forming the nucleosome particle. Previous studies demonstrated that histone tails are often subjected to post-translational modifications, including methylation, acetylation, ubiquitination, phosphorylation, and others. Most of these modifications are reversible. Taking the acetylation as an example, specific lysine residues of histone H3 and H4 are acetylated by the histone acetyltransferase (HAT) complexes, whereas the histone deacetylase (HDAC) complexes are responsible for removing these acetyl groups from histone tails. The acetylation level in eukaryotes is tightly controlled by HATs and HDACs. Abnormalities in acetylation lead to the occurrence of severe diseases and even cancers. Class I histone deacetylase enzymes, whose representatives are the Saccharomyces cerevisiae Rpd3S complex and its homolog Clr6S complex in Schizosaccharomyces pombe, utilize a zinc ion as its co-factor to catalyze the deacetylation. According to previous studies, Clr6S complex comprises six subunits, including Clr6, Pst2, Prw1, Alp13, Cph1, and Cph2, with a molecular weight of more than 400 kDa. Despite many years of study, high-resolution structural information on Rpd3S or Clr6S complex has not been reported.
The Laboratory of Structural Biochemistry at SIAIS has long been focusing on studying the mechanisms of transcriptional regulation and histone modifications in eukaryotes. Drs. Zhang Heqiao, Nie Yan and their colleagues reported the first near-atomic resolution cryo-EM structure of entire Mediator complex in 2021 in Molecular Cell (Zhang et al., Mol Cell 2021). In 2022, the group led by Prof. Roger Kornberg and Dr. Zhang Heqiao published a research article in PNAS, in which they reported the near-atomic resolution cryo-EM structure of the NuA4 histone acetyltransferase complex (Ji et al., PNAS 2022).
In the recently published work, the team successfully overexpressed the S. cerevisiae Rpd3S and S. pombe Clr6S complexes through insect-cell expression system, purified to homogeneity, set up a novel in vitro HDAC assay, and determined the structure of Clr6S at 3.2 Å through cryo-electron microscopy. The structure reveals, unexpectedly, two Alp13 subunits (an MRG-domain containing protein) in the complex. Two subunits Pst2 and Cph2 play scaffolding roles in the protein complex assembly. The structure also shows that Clr6, Pst2 and Prw1 form a core module independent of other subunits, among which the catalytic subunit Clr6 is embraced by scaffolding subunits Pst2 and Cph2, likely maintaining the active center in a right position and orientation for subsequent catalysis. The study also identified the catalytic zin ion, a water molecule serving as the nucleophile and significant residues important for catalysis, and therefore proposed a catalytic model. Intriguingly, a loop from Pst2, adjacent to the active center, is found essential for the catalytic activity of Clr6S, since deletion of this loop severely compromised the HDAC activity. The Pst2-loop (designated as “active center loop”) is likely important for substrate accommodation and improvement of the catalytic efficiency. The study also reported two cryo-EM maps of Rpd3S and Clr6S in complex with H3K36 methylation modified nucleosomes. The maps show that the nucleosomes bind to the concave sides of both two class I HDAC complexes, with multiple contacts involved.
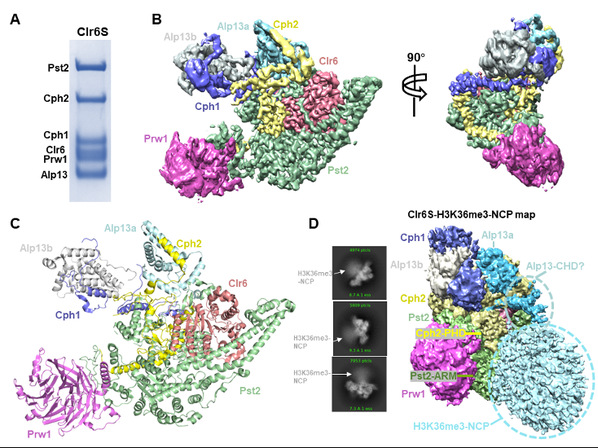
Purification and cryo-EM analysis of the class I histone deacetylase complex. (A) Purification of the Clr6S complex. (B) Cryo-EM map of the Clr6S complex. (C) Cryo-EM structure of the Clr6S complex. (D) Cryo-EM map of the Clr6S-H3K36me3-NCP
Third-year Ph.D. candidate Wang Xiao and senior engineer Wang Yannan from SIAIS are the co-first authors. Dr. Zhang Heqiao and Prof. Roger Kornberg are the co-corresponding authors. Dr. Liu Simiao from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Ms. Zhang Yi and Ph.D. candidate Xu Ke from SIAIS participated and made contributions to this project. ShanghaiTech University is the primary affiliation. All the cryo-EM data were collected at the Bio-Electron Microscopy Facility of ShanghaiTech University.
**This article is provided by the Laboratory of Structural Biochemistry

