Recently, a group led by Distinguished Adjunct Professor Roger Kornberg and Research Associate Professor Zhang Heqiao of the laboratory of structure biochemistry at SIAIS, published a paper entitled “Structure of the NuA4 histone acetyltransferase complex” in the Proceedings of the National Academy of the Sciences (PNAS), reporting a near-atomic resolution structure of the NuA4 complex in eukaryotes obtained by cryo-electron microscopy (cryo-EM).

The fundamental unit of eukaryotic chromosomes is the nucleosome, in which 147 base pairs of DNA are wrapped around an octamer of the histone proteins H2A, H2B, H3 and H4. Epigenetic phenomena arise, in part, from acetylation of the histones. There are two major histone acetyltransferase complexes in eukaryotes, SAGA and NuA4. SAGA is mainly responsible for acetylation of histone H3, whereas NuA4 mainly acetylates H2A and H4. Previous studies demonstrated that NuA4 plays important roles in the maintenance of genome stability and in transcriptional regulation. In S. cerevisiae, NuA4 is a 13-subunit complex, with a molecular weight of more than 1 MDa. Despite many years of study, high-resolution structural information on NuA4 complex has not been reported.
The laboratory of structure biochemistry has focused on transcriptional regulation in eukaryotes. Dr. Zhang and other researchers reported, in 2021 in Molecular Cell, the first near-atomic resolution structure of Mediator, the central molecule in transcriptional regulation.
The research team have now determined the cryo-EM structure of the endogenous NuA4 complex of S. cerevisiae at 3.8-4.0 Å resolution. The Eaf1 and Eaf2 subunits form the backbone of the structure, docking the other subunits to assemble the entire complex. In contrast with previous studies, the “neck” region of the structure is formed by Eaf1, Eaf2 and the amino-terminal domain of Epl1. Combining the cryo-EM structure and the AlphaFold-Multimer predicted structure revealed possible locations of the catalytic (Piccolo) module and the Yaf9 subunit of NuA4. Biochemical studies showed the Piccolo module and the complete NuA4 exhibit comparable histone acetyltransferase activities, but the Piccolo module alone interacts with nucleosomes much more strongly than the complete NuA4, indicating that other subunits of the NuA4 complex diminish the affinity of the Piccolo module for the nucleosome, enabling rapid movement from one nucleosome to another.
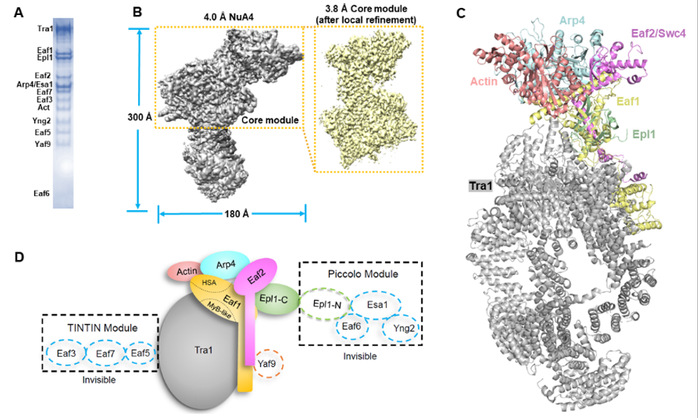
Structural studies of the NuA4 complex. (A) the purified NuA4 complex in this study. (B) the cryo-EM maps reported in this study. (C) the determined NuA4 structure. (D) a diagram illustrating the subunit composition and localization of NuA4 complex.
Ph.D. candidates Ji Liting and Xu Ke, and senior engineer Zhao Lixia from SIAIS are the co-first authors. Prof. Roger Kornberg and Dr. Zhang Heqiao are the co-corresponding authors. ShanghaiTech University is the primary affiliation. All the cryo-EM data were collected at the Bio-Electron Microscopy Facility of ShanghaiTech University.
*The news article is provided by the laboratory of structure of biochemistry

