On June 3, 2020, a research article entitled “Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response” was published online in Cell Reports. This work was done by a team led by Dr. Chen Jia from School of Life Science and Technology (SLST) at ShanghaiTech University, Dr. Yang Li from CAS-MPG Partner Institute for Computational Biology and Dr. Liu Zhen from Institute of Neuroscience & Center for Excellence in Brain Science and Intelligence Technology at Chinese Academy of Sciences.
The advent of base editors (BEs) holds great potential for correcting pathogenic-related point mutations to treat relevant diseases. However, Cas9 nickase (nCas9)-derived BEs lead to DNA double-strand breaks, which can trigger unwanted DNA damage response (DDR).
Based on the previously constructed dCas12a base editor, researchers screened a variety of cytosine deaminase (APOBEC/AID) and optimized hA3A-dcas12a-BE, which finally brought a new base editing system, BEACON (Base Editing induced by human APOBEC3A and Cas12a without DNA break). The BEACON system can accurately and efficiently mediate base editing while avoiding cell death caused by the activation of DNA damage response pathway.
In addition, the researchers applied the BEACON system in mice and successfully achieved accurate and safe base editing. BEACON is upgraded from two series of base editing systems, Cas12a-BE and hA3A-BE, and has the advantages of high efficiency, accuracy and safety, providing a better choice for the comprehensive application of the base editing system in basic research and future clinic.
In this study, researchers also developed a powerful RNA-editing analysis-pipeline to decode all-twelve-types of RNA-editing (RADAR, https://github.com/YangLab/RADAR). By using RADAR, researchers confirmed that BEACON only induced a background level of RNA C-to-U OT mutations, with the most common type of A-to-I RNA editing events in all cases.
This study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission, and research start-up funds of ShanghaiTech University.
Link to the paper: https://www.cell.com/cell-reports/fulltext/S2211-1247(20)30700-2
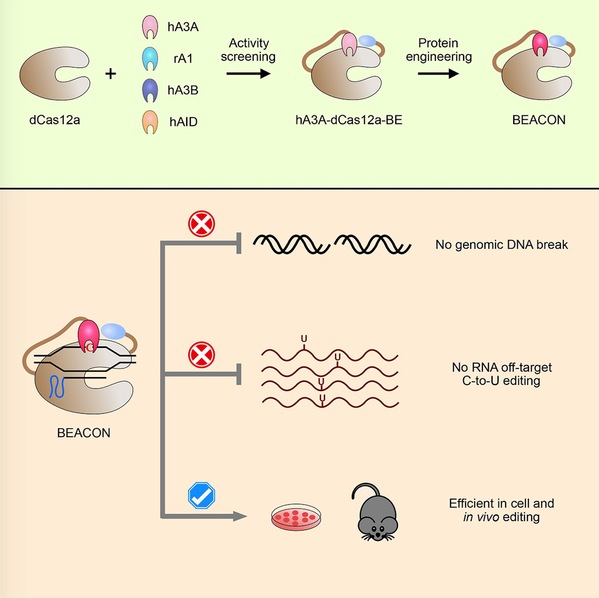
Figure 1. The development and applications of BEACON.

