Research teams led by SPST Assistant Professor Ji Quanjiang and Fudan University Professor Gan Jianhua recently used structural biology, biochemical, and cell-based genome editing experiments to reveal the mechanisms of the PAM expansion and fidelity enhancement of a new evolved Cas9 nuclease xCas9. Their work, “Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease”, was published online in the journal PLOS Biology.
CRISPR systems have been harnessed and engineered for robust genome editing in diverse areas, including medicine, biology, chemistry, agronomy and other fields. The most widely utilized Cas9 nuclease from Streptococcus pyogenes consists of a RNA-guided SpCas9 endonuclease and a single-guide RNA (sgRNA). The sgRNA forms a complex with the SpCas9 protein and directs the SpCas9 protein to the genomic DNA locus by base-pairing with the sequence that is adjacent to a PAM. The SpCas9/sgRNA complex creates a double-strand DNA break within the base-pairing region, stimulating the DNA repair pathway. Precise genome editing can be achieved through recombination of the target locus with exogenously supplied DNA “donor templates” during the process of homology-directed repair. However, the utility of SpCas9 in therapeutic and biological applications is restricted by its PAM compatibility as well as by its off-target effects.
The SpCas9 variant xCas9 recently evolved to possess broad PAM compatibility and substantially lower genome-wide off-target activity than WT SpCas9. However, the mechanism of why this evolved xCas9 protein possesses both high fidelity and wide PAM properties has not yet been clarified.
This work successfully solved the crystal structures of xCas9 in complex with sgRNA and DNA containing four different PAMs, and revealed that xCas9 adopts a unique two-mode PAM recognition mechanism. In addition, biochemical and cell-based genome editing experiments pinpointed the critical role of the key residues for PAM expansion and fidelity enhancement. This study provides critical insights into the mechanisms of the PAM expansion and fidelity enhancement of xCas9 and could further facilitate the engineering of SpCas9 and other Cas9 orthologs.
Dr. Chen Weizhong is the first author, and graduate student Zhang Hongyuan is the second author. Professors Ji Quanjiang and Gan Jianhua are the corresponding authors. This study was supported by Science Foundation of Ministry of Education, ‘Dynamic Modifications of Biomacromolecules and Chemical Intervention’ Major Research Plan, Shanghai Science and Technology Committee Rising-Star Program.
Read more at: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3000496
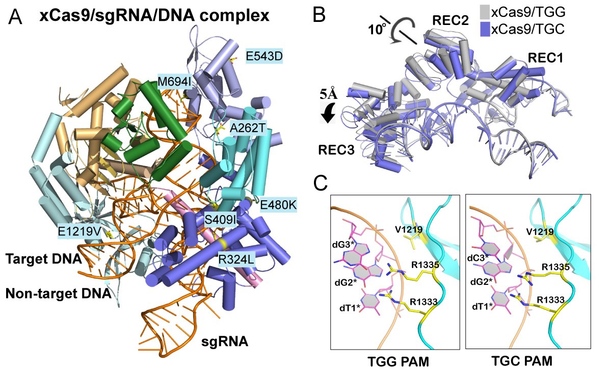
Figure A: Crystal structure of xCas9 in complex with sgRNA and targeted DNA. Figure B: xCas9 undergoes a notable structural rearrangement in the α-helix recognition domain (REC domain) when recognizing DNA sequences with different PAM sites. Figure C: When recognizing different PAM sequences, the key PAM-interacting residues of xCas9 exhibit significant conformational differences.

