Recently, a collaboration between the Ji Lab at the School of Physical Science and Technology, and Beijing Institute of Genomics’s Han Lab developed a single-base editing technique in Staphylococcus aureus and their results were published online in Chemical Science in an article titled, “Highly Efficient Base Editing in Staphylococcus aureus Using an Engineered CRISPR RNA-guided Cytidine Deaminase.”
As a major human pathogen, Staphylococcus aureus is the leading cause of hospital- and community-acquired infections. The pathogen can cause a wide variety of infections, including minor skin infections and life-threating diseases, such as endocarditis, necrotizing pneumonia, and toxic shock syndrome. The emergence of drug-resistant S. aureus, such as methicillin-resistant S. aureus (MRSA, a Superbug), has posed a severe public health crisis worldwide and has highlighted the urgent need to develop novel therapeutic means against drug-resistant S. aureus infections.
Before the development of the base-editing system, the Ji Lab had developed a CRISPR/Cas9-mediated genome editing system pCasSA (JACS, 2017, 139, 3790). The method allows for rapid and efficient genetic manipulation in S. aureus, accelerating bacterial physiology study, such as pathogenesis and drug resistance, and boosting novel drug-target exploration and new therapeutic method development. However, the weak intrinsic homologous recombination capacity of S. aureus prevents the high-rate recovery of survival cells after the double-stranded break of the genome, thus hampering the application of the pCasSA system in strains with low transformation efficiencies, such as many MRSA strains directly isolated from patients.
Ji Lab designed and constructed a highly efficient and convenient base-editing system pnCasSA-BEC in S. aureus via engineering a fusion of a Cas9 nickase (Streptococcus pyogenes Cas9D10A) and a cytidine deaminase (rat APOBEC1). The pnCasSA-BEC system can achieve efficient base editing without using repair templates or sacrificing transformation CFUs. In addition, the system enables efficient gene inactivation by converting the codons of CAA, CAG, CGA, and TGG to the stop codons of TAA, TAG, TGA, and TAA in the genomes. The development of the base editing system will dramatically accelerate study of bacterial physiology and drug-target exploration. In addition, it will provide critical insight into base-editing system development in other microbes.
Graduate student Gu Tongnian from the School of Physics and Science Technology of ShanghaiTech University, and Zhao Siqi from Beijing Institute of Genomics are the co-first authors of the paper and Professors Ji Quanjiang and Han Dali are corresponding authors.
Their research was supported by the National Key R&D Program of China, the National Natural Science Foundation of China. In addition, a patent application has been submitted for the technique.
Read more: http://pubs.rsc.org/en/content/articlelanding/2018/sc/c8sc00637g#!divAbstract
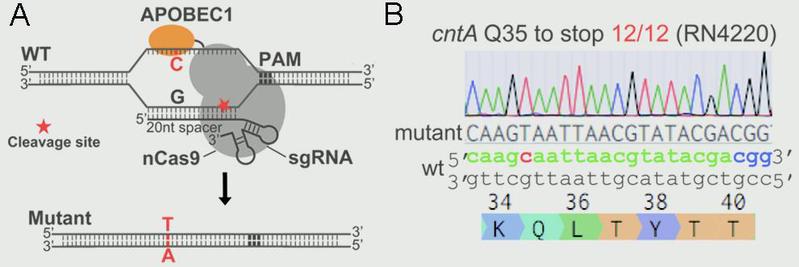
Figure 1.(A)Scheme of the editing mechanism of the “base editor” (pnCasSA-BEC). (B) The pnCasSA-BEC system enables highly efficient base editing in the cntA gene in the RN4220 strain.

