A team of scientists led by Dr. Jia Chen in the School of Life Science and Technology at ShanghaiTech University, Dr. Li Yang in the CAS-MPG Partner Institute for Computational Biology at the Chinese Academy of Sciences, and Dr. Bin Shen in the State Key Laboratory of Reproductive Medicine in the Department of Histology and Embryology at Nanjing Medical University has revealed that APOBEC3 induces mutations during the repair of CRISPR–Cas9-generated DNA breaks.
Members of the apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like or activation-induced cytidine deaminase (APOBEC-AID) family are single-strand-specific cytidine deaminases that are expressed ubiquitously in various cells and tissues and catalyze cytidine-to-uridine (C to U) base substitutions in RNA, viral DNA and genomic DNA. Previously, it has been found that the repair of preformed single-strand breaks (SSBs) can induce mutations in flanking DNA through a mechanism that involves the APOBEC3-induced cytidine deamination in single-stranded DNA (ssDNA). Interestingly, single-stranded nucleic acids are prevalent in CRISPR-Cas9-mediated gene editing processes. For instance, synthetic single-stranded oligodeoxynucleotides (ssODNs) can be used as homologous donors in homology-directed repair (HDR) for correcting mutations, and genomic ssDNA regions are also generated during the repair of DSBs and SSBs. The research team were intrigued to examine whether APOBEC3s can target these single-stranded nucleic acids to affect the repair outcome of CRISPR-Cas9-generated DNA breaks.
In their study, the researchers demonstrated that APOBEC3 can trigger cytidine deamination of ssODN, which ultimately results in base substitution mutations in genomic DNA through the homology-directed repair of Cas9-generated double-strand breaks. In addition, the APOBEC3-catalyzed deamination in genomic single-stranded DNA, formed during the repair of Cas9 nickase-generated single-strand breaks in human cells, can be further processed to yield mutations mainly involving insertions or deletions (indels). Both APOBEC3-mediated deamination and DNA-repair proteins play important roles in the generation of these indels.
In basic scientific research, although it is practical to sequence the edited genomic loci, and then select single clones without these unintended mutation byproducts, it is usually difficult to isolate single clones for postnatal gene correction of tissues or organs. Thus, the unintended mutations mediated by APOBEC3s should be avoided as much as possible when performing genome editing in tissues or organs, especially those with high APOBEC3 activity. This study showed that dsODN donors are largely resistant to the APOBEC-triggered deamination in Cas9-mediated HDR, suggesting the utilization of dsODN instead of ssODN as homologous donors for high-fidelity gene correction in postnatal tissues or organs. Also, temporarily suppressing endogenous APOBEC3s during the processes of gene editing can repress these unwanted mutations in genomic DNA.
Their paper, “APOBEC3 induces mutations during repair of CRISPR–Cas9-generated DNA breaks” was published online in Nature Structural & Molecular Biology on Dec 11, 2017. The study was supported by grants from the National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission, and ShanghaiTech University.
Read more at: https://www.nature.com/articles/s41594-017-0004-6
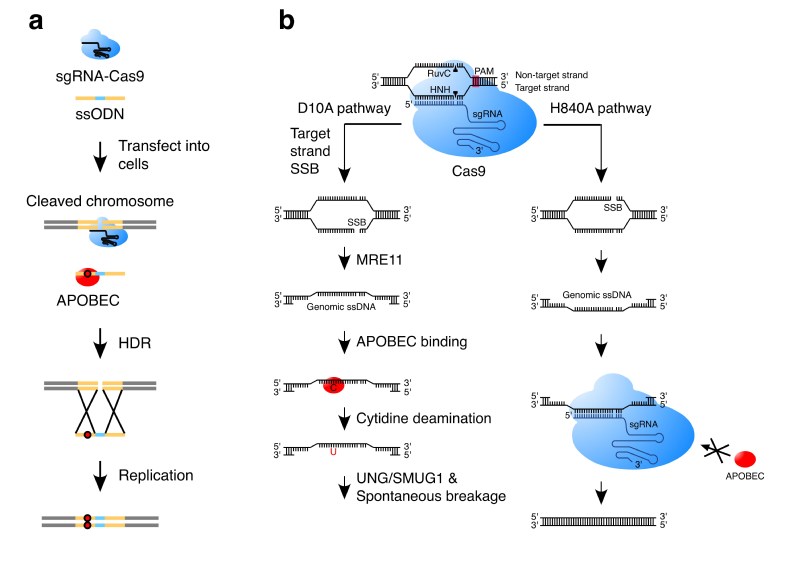
a. APOBEC induces base-substitution mutations during the HDR with an ssODN donor.
b. APOBEC induces indel mutations during the repair of single-strand breaks generated by Cas9 nickase D10A.

