Genome editing is a type of genetic engineering, which utilizes programmable nucleases such as the Cas9 nuclease to achieve desired base insertion, deletion or substitutions in the genome of living organisms. Directed by guide RNA (gRNA), Cas9 nuclease can generate DNA double strand breaks (DSBs) at targeted genomic sites. Subsequent resolving of the generated DSB through non-homologous end joining (NHEJ) DNA repair pathway would introduce random base insertions/deletions (indels) in the genome, thereby leading to open reading frame (ORF) shift and ultimately target gene inactivation. In contrast, activation of homology-directed repair (HDR) by the generated DSB allows the genomic DNA sequence at target site to be replaced by the sequence of exogenously provided donor DNA, thus resulting in base substitutions. In theory, CRISPR/Cas9-meditate HDR could be utilized to correct disease driven mutations and potentially cure genetic disease. In practice, however, the efficiency of HDR-mediated gene correction is low (normally < 5%) as NHEJ usually outcompetes HDR in resolving DSBs and because the occurrence of the latter is both cell type-specific and cell cycle-dependent. The relatively low efficiency of HDR thus limited the translation of CRISPR/Cas genome editing tools in the field of precision gene therapy.
To overcome the cell type and cell cycle dependency of HDR, base editors (BEs) were recently developed by combining the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like)/AID (activation-induced deaminase) cytidine deaminase family members with the CRISPR/Cas9 system to perform C-to-T base editing at targeted genomic sites. Mechanically, Cas9 variant-fused APOBEC/AID is directed to target sites by sgRNA, therefore introducing programmed C-to-T substitution at the single-base level. For maximum base editing efficiency, Cas9 nickase (nCas9) were utilized as genomic locators in the latest BE3. Nevertheless, non-negligible levels of indels (~4% to 12%) were observed in BE3-mediated base editing, though BE3 achieved much higher base substitution efficiency as compared to HDR. In addition, unwanted non-C-to-T (i.e., C-to-A or C-to-G) substitutions were also observed, the frequencies of which could be as high as that of desired C-to-T substitution in some examined cases. The existence of unwanted indels and C-to-A/C-to-G substitutions compromises the fidelity of base editing outcome.
A team of scientists led by Dr. Chen Jia at the School of Life Science and Technology, ShanghaiTech University, Dr. Li Yang at the CAS-MPG Partner Institute for Computational Biology, Chinese Academy of Sciences and Dr. Yang Bei at Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University has successfully overcome the relatively low fidelity of BE3 and developed an enhanced base editing system by co-expressing BE3 together with free UGI. This enhanced base editing system not only suppressed the formation of unwanted indels and base substitutions but also increased the frequency of C-to-T editing, thereby improving both the fidelity and efficiency of base editing. This finding holds great potential in extending the applications scope of base editor, especially in the translational field of precision medicine.
The paper entitled “Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor” was published online in Cell Research on August 29, 2017. This study was supported by the grants from National Natural Science Foundation of China, Ministry of Science and Technology, Shanghai Municipal Science and Technology Commission and ShanghaiTech University.
Read more at: http://www.nature.com/cr/journal/vaop/ncurrent/full/cr2017111a.html
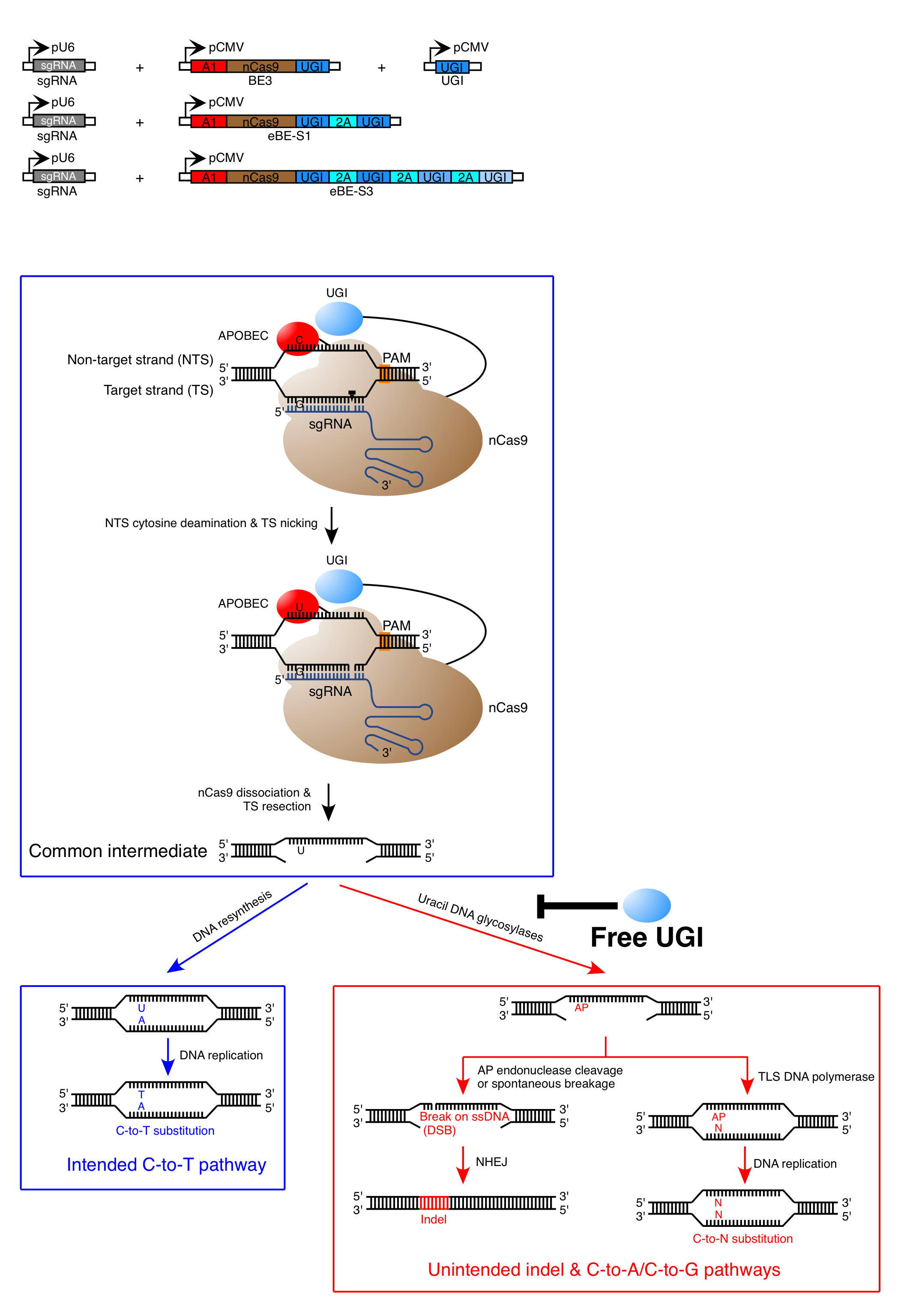
Enhanced Base Editing System and Mechanism of the Unintended Mutations Induced by BE3
- About
- News
- Faculty
- Research
-
Academics
- School of Physical Science and Technology (SPST)
- School of Life Science and Technology (SLST)
- School of Information Science and Technology (SIST)
- School of Entrepreneurship and Management (SEM)
- School of Creativity and Art (SCA)
- Institute of Humanities (IH)
- School of Biomedical Engineering (BME)
- Shanghai Institute for Advanced Immunochemical Studies (SIAIS)
- iHuman Institute
- Institute of Mathematical Sciences (IMS)
- Center for Transformative Science (CTS)
- Institute of Carbon Neutrality (ICN)
- Shanghai Clinical Research and Trial Center
- Tech Transfer
- Global
- Campus Life

