A team of scientists led by Dr. Huang Xingxu at the School of Life Science and Technology at ShanghaiTech and Dr. Liu Jiang at the Beijing Institute of Genomics, Chinese Academy of Sciences, has successfully described the chromatin interaction map of mice with male and female gametes and early embryos at different developmental stages and revealed global chromatin architecture reprogramming during embryogenesis by using small amounts of cell Hi-C technology. Their work entitled “3D chromatin structures of mature gametes and global structure reprogramming during embryogenesis in mammal” was published online in Cell on July 13, 2017.
Eukaryotic chromosomes contain a large amount of genetic information that is highly agglutinated and folded into an ordered 3D architecture in the nucleus to ensure that genes are correctly and orderly expressed in different cell types and developmental processes. Recent studies have revealed that the unit of chromatin 3D structure is a topologically associated domain (TAD) that is associated with histone modification, gene expression, and DNA replication. The TADs forming the basis for higher-level structures are called as ‘A’ and ‘B’ compartments. The ‘A’ compartment region tends to be active and the B compartment region tends to be inactive.
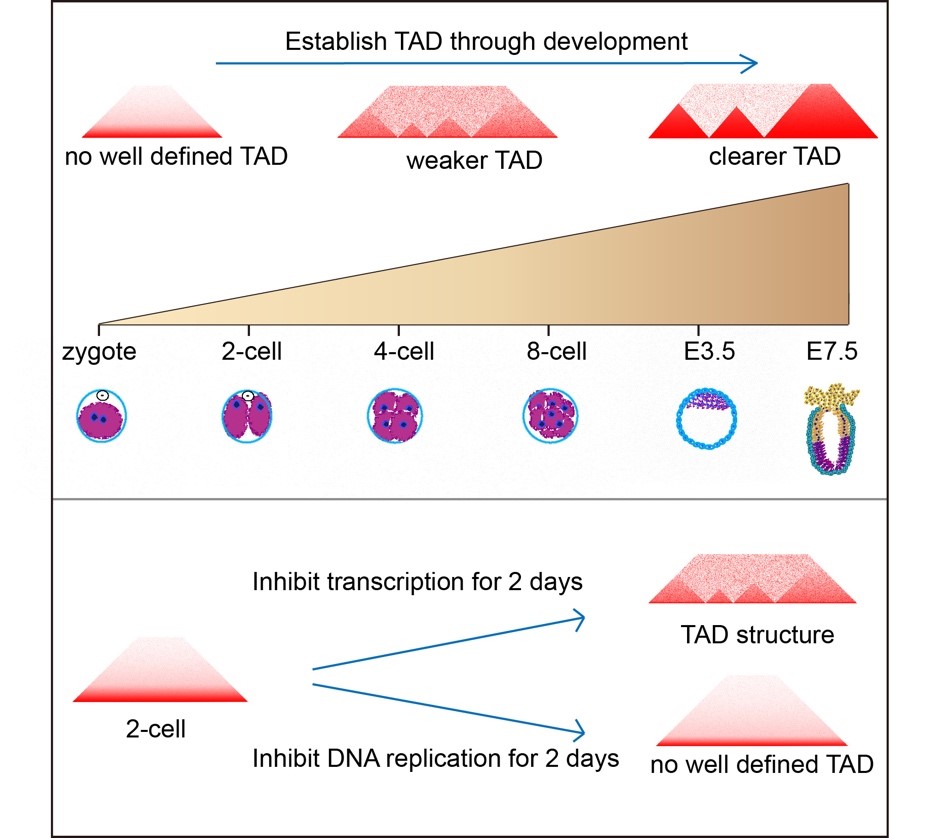
In mammals, mature oocytes poise at metaphase II stage, and sperm chromosomes are occupied by protamine, and the chromosomes of both male and female gametes are in a highly condensed state. This work shows that chromosomes of oocytes did not form the TAD structure and extra-long distance (>4 Mb) inter-TAD and interchromosomal interactions occur more frequently in sperms. After fertilization, the TAD structures of zygotes and 2-cell embryos are obscure, and the high-order chromosomal structure are gradually re-established during embryonic development. The study further showed that the establishment of TAD is dependent on DNA replication, but not the zygotic gene activation. This work also found that during the development of embryos CpGs demethylation occurred more frequently in the A component than the B component. In short, this study reveals that chromosome architecture processes a complex 3D structure reprograming during embryogenesis.
Dr. Xu Yanan and graduate student Feng Songjie are co-first authors and Dr. Huang Xingxu is the co-corresponding author. This work was supported by grants from CAS funding, The National Key R&D Program, the 973 Program of China, from National Natural Science Foundation of China.