A team of scientists led by Professor Liu Zhijie at ShanghaiTech’s iHuman Institute has determined the 3-dimensional molecular structure of the agonist-bound human cannabinoid receptor CB1. The work reveals the structural features of agonist-bound CB1 and the activation mechanism of the receptor. The results, described in a paper entitled "Crystal structures of agonist-bound human cannabinoid receptor CB1", were published online on July 6, 2017 in the prestigious journal Nature.
G protein-coupled receptors (GPCRs) are the largest and most diverse protein family in the cell membrane of the human body and play very important roles in signal transduction. About 40% of drugs on the market target GPCRs. The agonists will activate the receptor, while the antagonists will inhibit the activation of the receptor. Both agonists and antagonists show potential medication for drug discovery.
CB1 is one of the most highly expressed GPCRs in the human body and is the target of one of the longest used herbs, marijuana. Its ligands, including agonists and antagonists, can be used for treatment of pain, neurodegenerative disease, obesity, liver fibrosis. Last year, the iHuman team solved the 3D structure of CB1 in complex with antagonist AM6538 and their research was published in Cell. The work revealed the structural features of the antagonist-bound CB1and the binding modes of the antagonists, yet did not explain how agonists like THC, the major component of marijuana, bind to the receptor and how CB1 elicits its diverse physiological effects.
"Crystal structures of active state G protein-coupled receptor (GPCR) have been challenging to obtain due to their increased conformational flexibility and resulting instability," said Dr. Tian Hua, the first author of this paper. With a great deal of effort, the team finally solved the crystal structures of CB1 bound to the two novel agonists AM11542 and AM841 designed, synthesized and analyzed by Professor Alex Makriyannis' group at Northeastern University and characterized by Professor Laura Bohn's group at Scripps Florida.
"It is critical that we understand the different functional states of the receptors and their signal transduction pathways at atomic resolution", said Professor Raymond Stevens, Founding Director of the iHuman Institute. "We are thrilled to see the agonists AM11542 and AM841 could significantly stabilize the conformation of the receptor and lead to the structure determination. The two CB1-agonist complexes reveal significant conformational changes in the overall structure, relative to the antagonist-bound state, including a 53% reduction in volume of the ligand binding pocket and an increase in the surface area of the G protein binding region", said Professor Liu Zhijie, Vice Director of the iHuman Institute, the lead corresponding author of this paper.
"In addition, we observed a "twin toggle switch" of Phe2003.36/Trp3566.48 which appears to be essential in CB1 activation,” said Zhao Suwen, Research Associate Professor at iHuman Institute. “The structures also provide a molecular basis for predicting the binding modes of THC, the endogenous and synthetic cannabinoids and reveal important insights into the activation mechanism of CB1."
Other co-authors of this paper were Wu Yiran, Pu Mengchen, Ding Kang of iHuman Institute at ShanghaiTech University, Qu Lu from the Institute of Biophysics at Chinese Academy of Sciences, Kiran Vemuri, Spyros P. Nikas, Anisha Korde and Jiang Shan from Northeastern University, Gye Won Han at University of Southern California, Laura Bohn, Robert Laprairie and Jo-Hao Ho from The Scripps Research Institute, Florida, Liu Haiguang and Li Xuanxuan from Beijing Computational Science Research Center, and Michael Hanson of from GPCR Consortium.
This work was supported by the National Nature Science Foundation of China grant 31330019 (Z.J.L), the Ministry of Science and Technology of China grants 2014CB910400 (Z.J.L.) and 2015CB910104 (Z.J.L.), Natural Science Foundation of Shanghai 16ZR1448500 grant (S.Z.), National Key Research and Development Program of China grant 2016YCF0905902 (S.Z.), National Institutes of Health grants R01DA041435 (R.C.S., A.M), P01DA009158 (A.M., L.M.B.), R37DA023142 (A.M.) and NSF grants. Many thanks to the Shanghai Municipal Government and ShanghaiTech University for their financial support.
Read more at: http://dx.doi.org/10.1038/nature23272
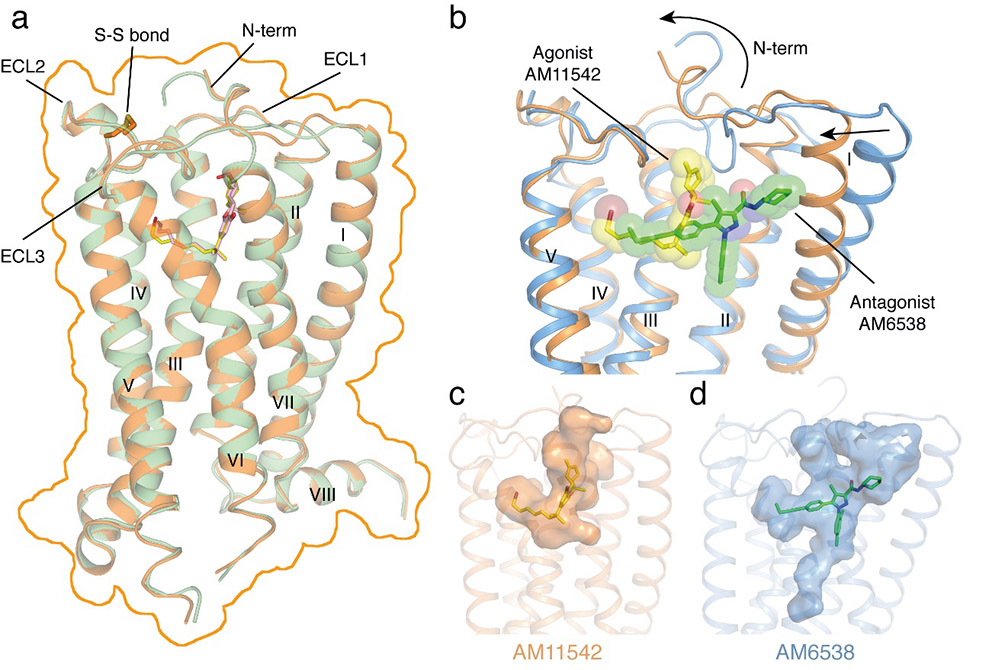
Figure 1: Overall structures of CB1–AM11542 and CB1–AM841 complexes and Agonist-bound (orange cartoon) and antagonist-bound (blue cartoon) CB1 ligand binding pockets. Agonist AM11542 (yellow) and antagonist AM6538 (green) are shown in sticks and sphere representations.

Figure 2:Professor Liu Zhijie’s research team at iHuman Institute
From left to right: Pu Mengchen, Wu Yiran, Liu Zhijie, Hua Tian, Raymond Stevens, Qu Lu, Ding Kang
- About
- News
- Faculty
- Research
-
Academics
- School of Physical Science and Technology (SPST)
- School of Life Science and Technology (SLST)
- School of Information Science and Technology (SIST)
- School of Entrepreneurship and Management (SEM)
- School of Creativity and Art (SCA)
- Institute of Humanities (IH)
- School of Biomedical Engineering (BME)
- Shanghai Institute for Advanced Immunochemical Studies (SIAIS)
- iHuman Institute
- Institute of Mathematical Sciences (IMS)
- Center for Transformative Science (CTS)
- Institute of Carbon Neutrality (ICN)
- Shanghai Clinical Research and Trial Center
- Tech Transfer
- Global
- Campus Life

