An international team led by the iHuman Institute, ShanghaiTech University has determined the 3-dimensional molecular structure of the human glucagon-like peptide-1 receptor (GLP-1R) drug binding domain. The work reveals molecular mechanisms of allosteric regulation in class B G protein-coupled receptors (GPCRs). The results, described in a paper entitled “Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators,” was published online on May 17, 2017 in the journal Nature. This paper is published simultaneously with a companion paper led by colleagues at the Shanghai Institute of Materia Medica (SIMM) describing the full-length glucagon receptorin the same journal.
Secreted by the gut, glucagon-like peptide-1 (GLP-1) exerts its glucose control on the human body by binding to GLP-1R that is expressed on the surfaces of specific cells (e.g.,pancreatic β cells). GLP-1R is a well-recognized drug target for type 2 diabetes, exemplified by several peptidic therapeutic agents on the market, with combined sales of several billions of dollars each year. Although effective, the peptide therapeutics are not orally available and many have side effects. Worldwide, the costs associated with treating diabetes and its complications are estimated to exceed $200bn a year. The number of patients with diabetes is growing at an alarming rate throughout the world, with the most significant and recent growth occurring in China.
The high resolution GLP-1R structure was determined in complex with two negative allosteric modulators (NAMs), respectively. Both structures are in an inactive conformation with each NAM bound in a similar pocket but distinctly different binding modes on the external surface of the receptor. Molecular modeling and mutation analysis suggest that agonist positive allosteric modulators (PAMs) target the same general region, but in a distinct sub-pocket.While the NAMs block the activation of GLP-1R by inserting into the cavity between helices VI and VII, PAMs binds mainly to the space between helices V and VI enabling the activation. “This structure is one of the holy grails of GPCR drug discovery” said Professor Raymond Stevens who co-led the study at the iHuman Institute, ShanghaiTech University.
Since most peptide drugs are administrated via non-oral routes, orally available GLP-1 or its small molecule surrogates have been vigorously sought after by many of the multi-national pharmaceutical companies. Determination of the three-dimensional structure of GLP-1R is thus helpful to better design and develop new therapeutics targeting the receptor. “GPCRs that bind peptides can be particularly challenging for small molecule drug discovery given multiple points of connection between the peptide and receptor. With the atomic resolution data, we can now see the atoms in the binding site and design safer next generation small molecule therapeutics” said iHuman Research Associate Professor Gaojie Song.
"This major undertaking started in 2002 when we were looking for small molecule GLP-1R agonists." said Professor Ming-Wei Wang of Shanghai Institute of Materia Medica and Fudan University who co-led the study.
"Our failed efforts in making the first orally active GLP-1R agonist Boc5 led us to conclude that high resolution structural biology is the preferred solution to druggability. Our research collaboration with the Stevens laboratory in Shanghai has been extremely rewarding and productive. We have made several significant discoveries that are impacting and will continue to impact the drug discovery field for many years to come. In the end, the biggest winner will be the patients ill with metabolic disorders."
The team effort was also co-led by iHuman Institute Professor and Deputy Director Zhi-Jie Liu. Other contributors in this study include Yuxia Wang, Qingtong Zhou, Kaiwen Liu, Dongsheng Liu, Suwen Zhao, Yiran Wu, and Fan Wu from ShanghaiTech University, Dehua Yang, Xiaoqing Cai, Antai Dai, Guangyai Lin, and Beili Wu from Shanghai Institute of Materia Medica, Shanshan Jiang and Li Ye from Fudan University, Chris de Graaf from Vrije Universiteit Amsterdam, Gye Won Han from University of Southern California, Jesper Lau from Novo Nordisk, and Michael A. Hanson from GPCR Consortium. Financial support for this work comes from, in part, Shanghai Municipal Government, ShanghaiTech University, Shanghai "Pujiang Talents" grant, GPCR Consortium, National Natural Science Foundation of China, National Health and Family Planning Commission, Ministry of Science and Technology of China, Chinese Academy of Sciences, and the Shanghai Science and Technology Development Fund.
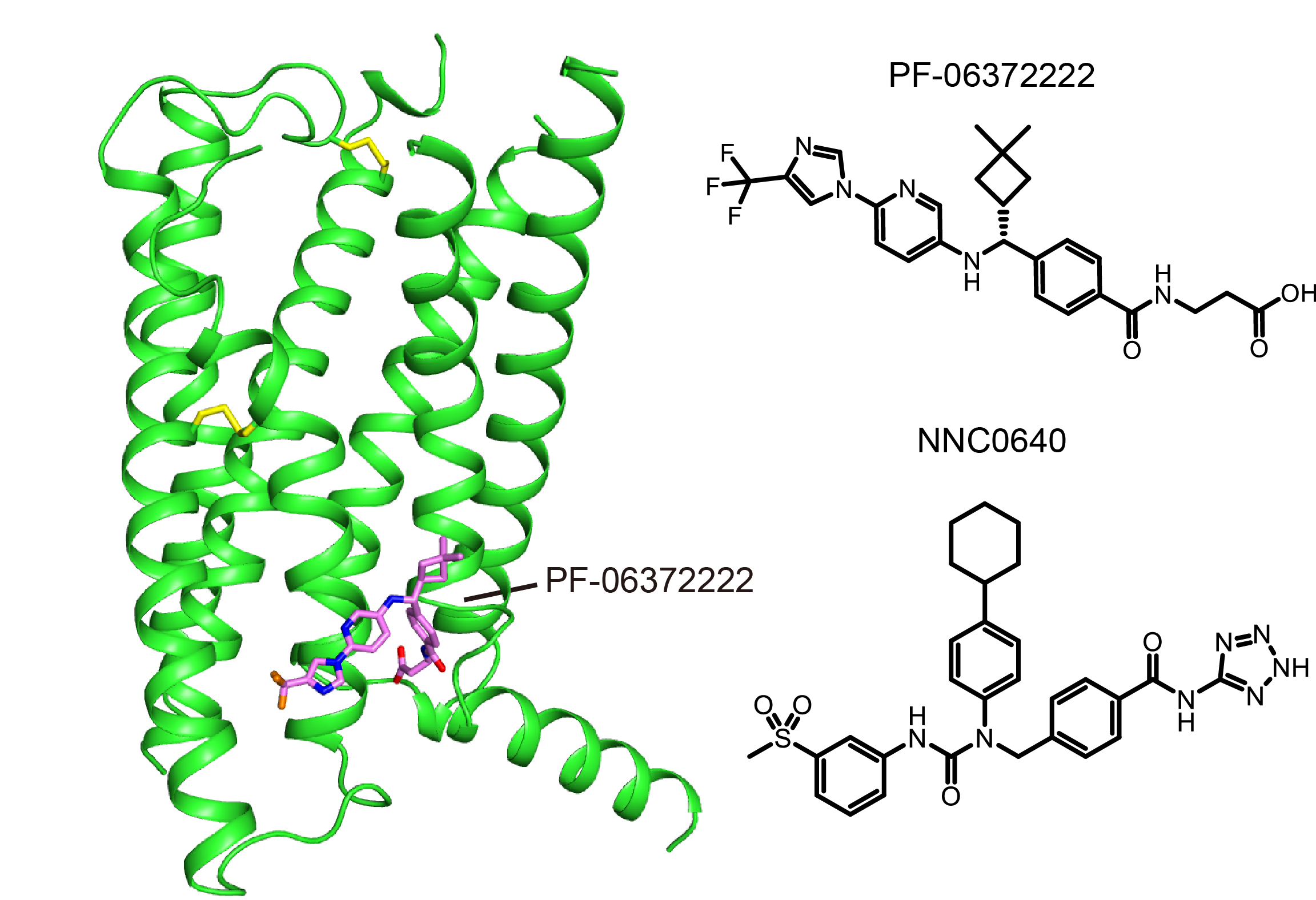
Crystal structure of GLP-1R bound to 2 different NAMs

From left to right: Qingtong Zhou ,Yiran Wu, Kaiwen Liu, Zhi-Jie Liu, Yuxia Wang, Gaojie Song, Raymond Stevens, Wu Fan, Suwen Zhao, Dongsheng Liu.

