A team of scientists led by Prof. Chen Jia at the School of Life Science and Technology (SLST) of ShanghaiTech University, Prof. Yang Bei at Shanghai Institute of Advanced Immunochemical Studies (SIAIS) of ShanghaiTech University, Prof. Yang Li at the Shanghai Institute of Nutrition and Health of Chinese Academy of Sciences and Prof. Yin Hao at Wuhan University published a technical report article entitled “Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations” in Nature Cell Biology on May 10, 2021. The research team successfully used a cleavable deoxycytidine deaminase inhibitor (dCDI) domain to construct a transformer base editor (tBE) system that induces efficient editing with only background levels of genome-wide and transcriptome-wide off-target mutations. They also delivered the tBE system into mice through a dual-adeno-associated virus (AAV) system, which created a premature stop codon in Pcsk9 and significantly reduced serum PCSK9, resulting in a ~30–40% decrease in total cholesterol.
The fusion of CRISPR–Cas9 with cytidine deaminases produces base editors (BEs) capable of programmable C-to-T editing, which has potential in clinical applications but suffers from off-target mutations. According to ClinVar database, BEs could correct more than half of known human pathogenic point mutations. However, recent studies revealed that BEs could induce both genome-wide and transcriptome-wide off-target mutations. Importantly, these off-target mutations occurred at the sites with no sequence similarity to sgRNA and thus cannot be predicted, which impedes BE’s applications, particularly for therapeutics.
In this study, researchers first developed a quantitative method by co-expressing S. aureus and S. pyogenes Cas9 orthologs (CESSCO) to detect the mutations induced by the APOBEC moiety of BEs at sgRNA/Cas9-independent single-stranded off-target sites. Next, they identified some deoxycytidine deaminase inhibitor (dCDI) domains and took advantage of mA3dCDI domains and split-TEV system to develop a novel tBE system. The tBE system remains inactive at off-target sites with a cleavable fusion of dCDI domain, thus eliminating unintended mutations. Only when binding at on-target sites, tBE is transformed to cleave off the dCDI domain and catalyzes targeted deamination for precise editing (Figure 1). Adeno-associated virus (AAV) is an efficient and widely used delivery agent, which has a genome packaging size limit of ≤4.7 kb. The size of the traditional BEs precludes packaging in a single AAV vector. However, the tBE system breaks from regular APOBEC-Cas9 backbone of BE, which could be conveniently packaged into a dual-AAV system to maintain editing efficiency maximally. As editing precision is essential for BEs, the tBE system developed by the team for eliminating off-target mutations sheds light on therapy-related applications.
Dr. Wang Lijie, along with PhD candidate Gao Runze from Chen Jia’s Lab, PhD candidate Xue Wei from Yang Li’s Lab, postdoctoral scholar Zhang Hongxia, and PhD candidate Qiu Houyuan from Yin Hao’s Lab are the co-first authors. Prof. Chen Jia, Prof. Yang Bei, Prof. Yang Li and Prof. Yin Hao are the co-corresponding authors.
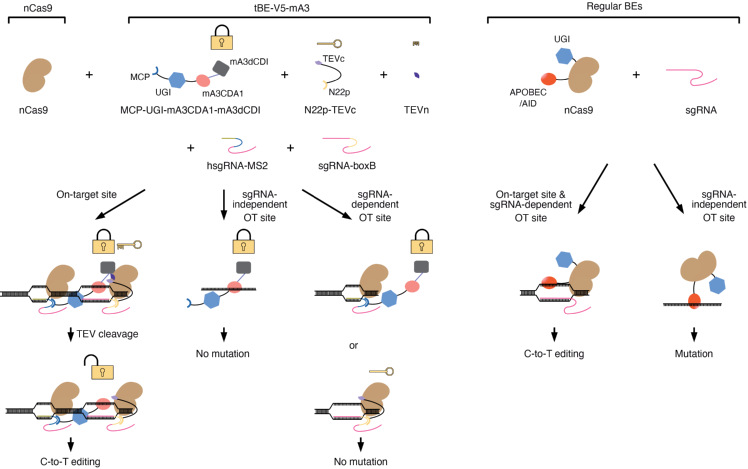
Figure 1. Schematic illustrating that tBE-V5–mA3 induces base editing at on-target, but not sgRNA-independent or sgRNA-dependent off-target sites.
Link of the article: https://www.nature.com/articles/s41556-021-00671-4

