In a review article entitled “A Tale of Two Moieties: Rapidly-evolving CRISPR/Cas-based Genome Editing”, Prof. Chen Jia at School of Life Science and Technology (SLST) of ShanghaiTech University and Prof. Yang Li at CAS-MPG Partner Institute for Computational Biology (PICB) of Chinese Academy of Sciences, summarized recent progress about the development and application of gene editing tools. The review article was published online in Trends in Biochemical Science on June 30, 2020.
The completion of the human genome project in the beginning of this century and the application of affordable high-throughput sequencing technologies in the past decade have led life science research to the post-genome era with genome-wide understanding of functional genomic elements related to human health and diseases. Importantly, the advent of practical genome editing technologies provides powerful methods to change genetic information, which benefits not only basic research aiming to decipher how different genotypes result in distinct phenotypes, but also pre-clinical study to cure human diseases caused by genetic mutations.
Two major moieties in genome editing are required for precise genetic changes: the locator moiety for target binding and the effector moiety for genetic engineering. By taking advantage of CRISPR/Cas that consists of different modules for independent target binding and cleavage, a spectrum of precise and versatile genome editing technologies have been developed for broad applications in biomedical research, biotechnology and therapeutics. In this review, Prof. Chen Jia and Prof. Yang Li summarized the progress of genome editing systems from a view of both locator and effector moieties, and highlight the advance of newly reported CRISPR-conjugated base editing and prime editing systems. The authors also underscore distinct mechanisms of off-target effects in CRISPR-conjugated systems, and also discuss possible strategies to reduce off-target in the future.
Paper Link:
https://www.cell.com/trends/biochemical-sciences/fulltext/S0968-0004(20)30150-X
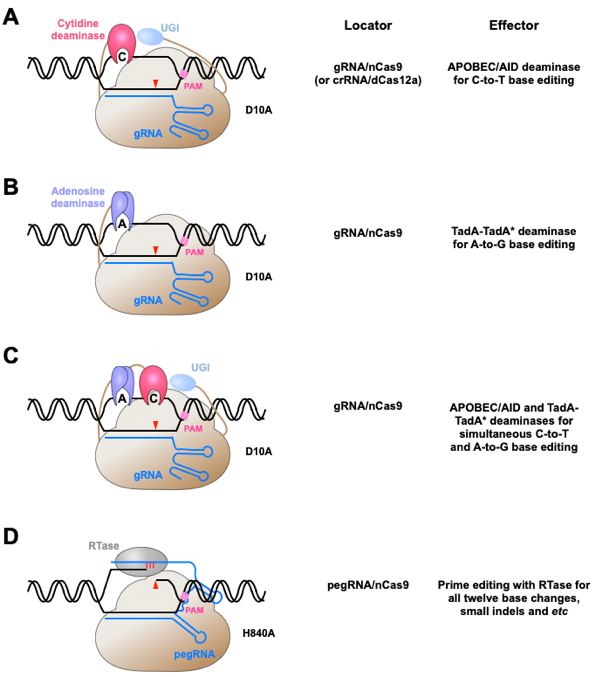
Figure 1. Conjugating nucleobase deaminases or reverse transcriptases with CRISPR/Cas proteins to achieve precise genome editing at single nucleotide resolution.
(A), (B) Schematic drawing of CBE (A) and ABE (B). Single base change of cytosine or adenine has been achieved by fusing cytidine (A) or adenosine (B) deaminase with Cas9 nickase (D10A). Of note, uracil DNA glycosylase inhibitor (UGI) is included in CBEs, but not ABEs, to reduce the formation of unwanted indels by CBEs. (C) Schematic drawing of dual function BE for simultaneous cytosine and adenine deamination. (D) Schematic drawing of PE. The conjugation of reverse transcriptase (RTase) with Cas9 nickase (H840A) leads to a versatile PE system for all type of base substitutions, small indels and their combinations. pegRNA, prime editing guide RNA.

