Groups of professors Liu Ji-Long,Zhuang Min, and Lu Pengfei from the School of Life Science and Technology (SLST) at ShanghaiTech University have jointly developed a new method CRUIS, or CRISPR-based RNA-United Interacting System, for capturing RNA-protein interactions on specific RNAs. It is a hybrid of two molecular machineries, CRISPR-based RNA-targeting Cas nuclease (dCas13a) and a proximity-labeling system (PUP-IT). The related work was published online on March 6th, 2020 under the title "Capturing RNA-protein interactions via CRUIS" in Nucleic Acids Research.
RNA-binding proteins (RBPs) play important roles in various biological processes such as variable RNA splicing, subcellular localization, translation, and stability. RNA and RBPs mutually regulate each other's behavior and functions, and thus are essential for various fundamental aspects of cell behavior. In order to gain insight into the function of an RNA and RBPs, it is necessary to identify detailed interactions between the RNA and its binding proteins. Traditional methods, including RIP and CLIP technology, for studying RNA-protein interactions couldn’t capture RNA-protein interactions of a specific RNA under physiological conditions in living cells. Thus, a huge advantage of CRUIS, as an RNA-centric method, is its ability to target a specific RNA and label its RBPs under physiological conditions of living cells. The authors have recently filled a PCT application for the CRUIS-based technologies.
In this study, the researchers combined a CRISPR-based RNA-targeting Cas nuclease and proximity-labeling system (PUP-IT), which used dCas13a as a tracker to target specific RNA sequences, while the proximity enzyme PafA is fused to dCas13a to label the surrounding RNA-binding proteins. Subsequently, the labeled proteins are enriched and identified by mass spectrometry. This project was benefited from technical support from the Mass Spectrometry Core Facility and the Molecular Imaging Core Facility at SLST. PhD candidates Zhang Ziheng, Sun Weiping, and Shi Tiezhu are the co-first authors, while professors Liu Ji-Long, Min Zhuang, and Lu Pengfei are the co-corresponding authors. The research was conducted at ShanghaiTech University.
Link to the paper: https://doi.org/10.1093/nar/gkaa143
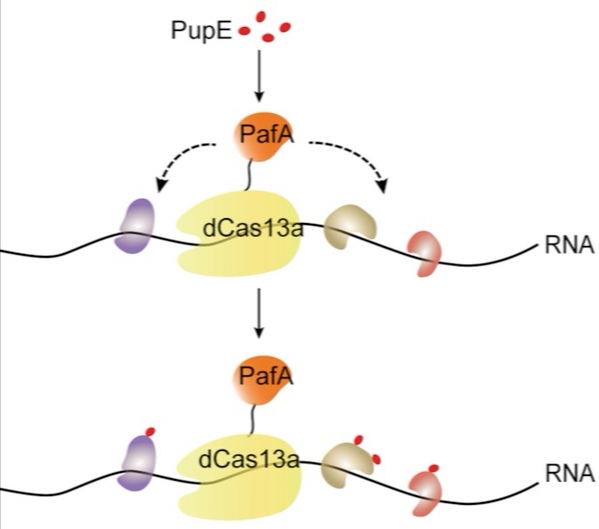
Figure 1 Design of CRUIS, a hybrid of CRISPR (yellow) and PUP-IT (orange)

