A research group led by iHuman Institute and SLST Assistant Professor Zhao Suwen, working with Professor Wang Ming-Wei from Shanghai Institute of Materia Medica/The National Center for Drug Screening/Fudan University and Professor Babu Madan from MRC Laboratory of Molecular Biology, recently discovered a common activation pathway in class A GPCRs. Their study, “Common activation mechanism of class A GPCRs, was published in the journal eLife on December 19, 2019.
G protein-coupled receptors (GPCRs) influence virtually every aspect of human physiology and are important drug targets with 475 marketed drugs (~34% of all FDA approved therapeutic agent agents). GPCR activation is an allosteric process transducing various external stimuli into cellular responses agonist binding induced G protein recruitment. Understanding the activation mechanism of GPCR is of paramount importance in pharmacology research and drug discovery. It is well established that outward movement of transmembrane helix 6 (TM6) upon ligand binding is a common feature of receptor activation, but the residue level changes that trigger the movement of TM6 remain less understood (Fig. 1).

Fig 1. Common GPCR activation mechanism and the residue level triggers are not well understood.
Using a residue-residue contact score (RRCS)-based framework, Professor Zhao and colleagues quantified the global, local, major and subtle conformational changes in a systematic way (i.e., inter-helical and intra-helical, switching and repacking contacts). By analyzing the conformational changes in 234 structures from 45 class A GPCRs, they discovered a common GPCR activation pathway comprising of 34 residue pairs and 35 residues (Fig. 2). The pathway unifies previous findings into a common activation mechanism and strings together the scattered key motifs such as CWxP, DRY, Na+ pocket, NPxxY and PIF, thereby directly linking the bottom of ligand-binding pocket with G protein coupling region. Collectively, the intra-helical/inter-helical and switching/repacking contacts between residues is not only critical to reveal the continuous and modular nature of the activation pathway, but also to link residue-level changes to transmembrane helix-level changes in the receptor.
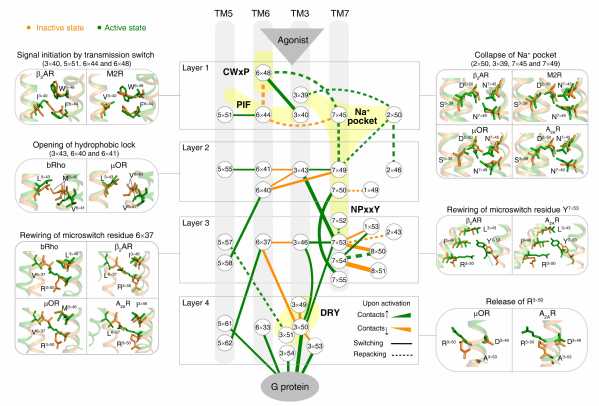
Fig 2. Universal activation pathway of class A GPCRs.
Site-directed mutagenesis experiments support this observation and reveal that rational mutations of residues in this pathway can be used to obtain receptors that are constitutively active or inactive. The common activation pathway provides the mechanistic interpretation of constitutively activating, inactivating and disease mutations. As a module responsible for activation, the common pathway allows for decoupling of the evolution of the ligand binding site and G protein binding region. The researchers suggest that “such an architecture might have facilitated GPCRs to emerge as a highly successful family of proteins for signal transduction in nature.”
ShanghaiTech University Research Assistant Professor Zhou Qingtong from Zhao Suwen’s group and Shanghai Institute of Materia Medica Professor Yang Dehua from Ming-Wei Wang’s group are the first authors. This work was supported by grants from National Natural Science Foundation of China, National Key R&D Program of China, National Mega R&D Program for Drug Discovery, Shanghai Science & Technology Development Fund, Novo Nordisk-CAS Research Fund, the Medical Research Council, National Institutes of Health Grants and annual overhead support from ShanghaiTech University and Chinese Academy of Sciences.
Read more at:https://elifesciences.org/articles/50279

