Recently, a research group led by SPST’s Dr. Ji Quanjiang established an efficient genome engineering platform in Acinetobacter baumannii firstly. Their research was published in Cell Chemical Biology with the title of “A highly efficient CRISPR-Cas9-based genome engineering platform in Acinetobacter baumannii to understand the H2O2-sensing mechanism of OxyR”.
This platform allows for rapid and precise gene deletion, insertion, point mutation, and C→T conversions in various strains of A. baumannii. By using these editing methods, the research group explored the new mechanisms that play critical roles in H2O2 sensing of OxyR and interpreted the detailed imipenem and sulbactam resistance mechanisms in the clinically isolated A. baumannii strain XH386.

Acinetobacter baumannii is one of the most threatening Gram-negative human pathogens and responsible for a wide variety of infectious diseases. In recent years, the progressing emergence of infections caused by multidrug-resistant (MDR) or extensively drug-resistant (XDR) A. baumannii has posed severe threats to public health worldwide. In 2017, the world health organization (WHO) categorized A. baumanni as the top-ranked critical pathogen, which emphasizes desperate needs for novel therapeutic strategies against A. baumannii infections. New therapeutic-means development requires a comprehensive understanding of bacterial pathogenesis and drug-resistance mechanisms. However, the current genetic manipulation methods which can achieve this goal are time-consuming and inefficient, such as the non-replicative plasmid-based allelic exchange method and the recombinase-catalyzed homologous recombination method.
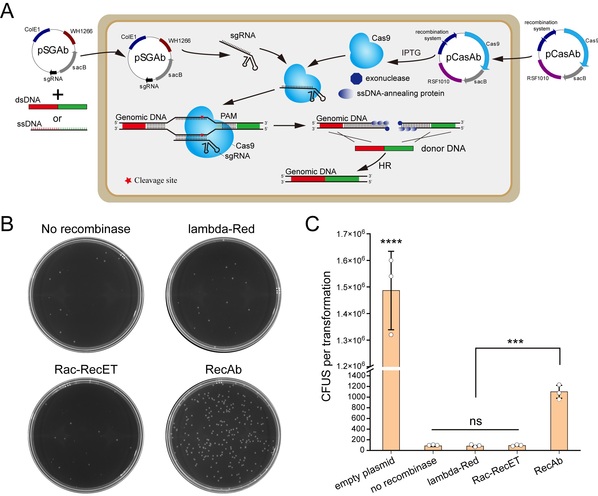
Figure 1. The pCasAb-pSGAb/RecAb genome editing platform in Acinetobacter baumannii. (A) Procedure scheme of pCasAb-pSGAb/RecAb system. (B, C) Only the RecAb recombination system can repair the DSB generated by CRISPR-Cas9 in A. baumannii.
In this study, researchers discovered that only the RecAb recombination system from A. baumannii IS-123 strain could repair the double-stranded DNA break generated by the CRISPR-Cas9 system in A. baumannii rather than the widely utilized E. coli phage RecET or λ-Red system. By coupling the CRISPR-Cas9 genome cleavage system with the A. baumannii RecAb recombination system, researchers developed the two-plasmid pCasAb-pSGAb/RecAb genome editing system to allow for efficient and precise gene deletion and insertion in diverse A. baumannii strains. Based on the two-plasmid system, a two-step insertion-deletion (In-Del) strategy for rapid single-nucleotide mutations in the genomic loci that lack an editable PAM site of CRISPR-Cas9 was also designed. With the use of the In-Del strategy, the researchers performed alanine scanning mutagenesis of 13 residues residing in the H2O2-sensing pocket to dissect the oxidative stress-sensing mechanism of OxyR.
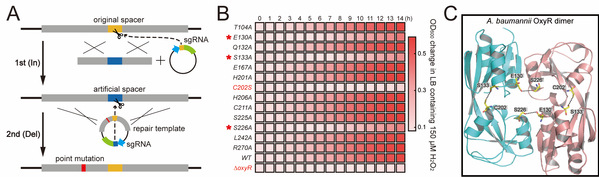
Figure 2. Dissection of the oxidative stress-sensing mechanism of OxyR using the In-Del strategy. (A) Procedure scheme of In-Del strategy. (B) Alanine scanning mutagenesis of residues residing in OxyR active pocket using In-Del strategy. (C) The new ROS-sensing mechanism of OxyR.
In order to simplify the gene-disruption processes furtherly, researchers adopted the fusion of the cytidine deaminase APOBEC1 and the Cas9 (D10A) nickase to develop a single-plasmid cytidine base-editing system pBECAb, allowing for efficient C to T conversion in a programmable manner in A. baumannii. The pBECAb system is capable of inactivating target genes by converting any of the four codons CAA, CAG, CGA, and TGG) into premature stop codons (TAA, TAG, and TGA). Exploiting this powerful technique, researchers systematically investigated the resistance mechanisms of imipenem and sulbactam in a clinically isolated multidrug resistant A. baumannii strain XH386 and unveiled the distinct roles of these genes in drug resistance through generating premature stop codons in three possible β-lactamase genes.
Postdoc Wang Yu is the first author, graduate student Wang Zhipeng is the second author, and Assistant Professor Ji Quanjiang is the corresponding author. This study was assisted by the research group of Professor Yu Yunsong from Sir Run Run Shaw Hospital at Zhejiang University. Two patents (201910644444.1 and 201910644324.1) about the related research results of this paper were submitted. This study was supported by ‘Dynamic Modifications of Biomacromolecules and Chemical Intervention’ Major Research Plan and Shanghai Science and Technology Committee Rising-Star Program.
It's worth mentioning that Ji group has developed several efficient genome editing tools in recent years, which could be applied to various major human pathogens, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Klebsiella pneumonia. With the use of the editing tools and the structural biology methods, the researchers conducted the further study on the pathogenesis and drug resistance mechanisms of pathogens, especially the mechanism of metal acquisition. All the genome editing tools developed by Ji group were shared on the Addgene website (https://www.addgene.org/Quanjiang_Ji/).
Read more at: https://www.cell.com/cell-chemical-biology/fulltext/S2451-9456(19)30277-6

